Examining the Negative Impact of Enzymes on Tissue Dissociation
Tissue dissociation is a valuable technique that enables researchers to isolate individual cells or smaller tissue fragments from complex tissues for downstream applications such as single-cell RNA sequencing (scRNA-seq) or fluorescent activated cell sorting (FACS).
In scRNA-seq, generating heterogeneous cell populations through tissue dissociation enables researchers to analyze gene expression profiles, identify distinct cell types, uncover cellular heterogeneity, and understand cellular diversity within tissues.
Immunotherapy is another application that often requires tissue be dissociated into heterogeneous immune cell populations. This enables researchers to study immune responses, immune cell interactions, and develop immunotherapeutic strategies. The isolation and characterization of different subsets of immune cells through FACS can aid in the design of targeted immunotherapies against cancer or autoimmune diseases. This requires the retention of the native cell surface markers through the tissue dissociation process.
A popular method used by many researchers to dissociate tissue is enzymatic tissue dissociation. Enzymatic tissue dissociation involves the use of proteolytic enzymes such as collagenase, trypsin, and dispase to break down the extracellular matrix (ECM) surrounding cells. While these enzymes are effective in breaking down the ECM, they can also damage cells in the process. While enzymatic tissue dissociation is widely used due to its efficiency and preservation of cell viability, the use of enzymes in tissue dissociation also poses potential negative impacts.
Enzymatic tissue dissociation raises concerns for many scientists as experiments have demonstrated the potential loss of cell functionality and alterations in cellular heterogeneity as result of enzymatic dissociation. Enzymes not only degrade the ECM but also affect the surrounding microenvironment and signaling molecules crucial for cell behavior and function. Enzymatic treatment may disrupt important cell-cell interactions, cell-matrix interactions, and signaling pathways, leading to changes in cellular behavior and functional properties.
Shen C, et al demonstrated the loss or alteration of certain cell surface markers, specifically lower CD27 expression in tissue dissociated with collagenase1. Additionally, Autengruber, et al demonstrated immune cell digestion with high dispase concentrations dramatically decreased surface expression of CD4, CD8, CD19, CD162, CD62L, CD25, CD27, ICOS-L, and PD-1L2 . Mattei D et al demonstrated the risk of introducing technical biases and biological artifacts when implementing enzymatic digestion-based isolation methods on transcriptional and proteotype profiles in mouse brain tissue by taking into account single-cell transcriptomics of brain cells, proteotypes of microglia and astrocytes, and flow cytometric analysis of microglia3. The use of enzymes in tissue dissociation also revealed induction of stress response genes through bulk RNA seq profiling of single-cell suspensions4.
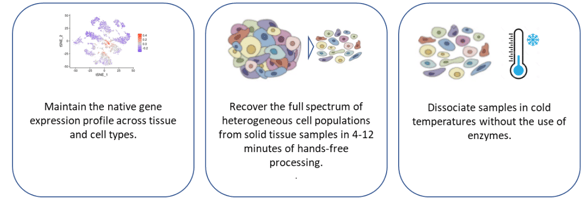
While popular, the use of enzymatic tissue dissociation has documented drawbacks which may impact the underlying cell functionality that is being studied. Scientists are seeking new technologies to streamline their tissue dissociation while not altering the biology of the cells they need to isolate. An alternative to the use of enzymes and heat is the Bulk Lateral Ultrasonic (BLU) energy used in the SimpleFlowTM platform from Cellsonics. BLU gently disaggregates solid tissue through gentle ultrasonic sound without applying heat or enzymes. This fast, automated process dissociates tumors or tissue into a single cell suspension in as little as 3 minutes per sample while retaining the sample at a constant temperature of 6oC.
References
- Shen C, Xu H, Alvarez X, Lackner AA, Veazey RS, et al. (2015) Reduced Expression of CD27 by Collagenase Treatment: Implications for Interpreting B Cell Data in Tissues. PLOS ONE 10(3): e0116667. https://doi.org/10.1371/journal.pone.0116667
- Autengruber A, Gereke M, Hansen G, Hennig C, Bruder D. Impact of enzymatic tissue disintegration on the level of surface molecule expression and immune cell function. Eur J Microbiol Immunol (Bp). 2012 Jun;2(2):112-20. doi: 10.1556/EuJMI.2.2012.2.3. Epub 2012 Jun 13. PMID: 24672679; PMCID: PMC3956959.
- Mattei D, Ivanov A, van Oostrum M, Pantelyushin S, Richetto J, Mueller F, Beffinger M, Schellhammer L, Vom Berg J, Wollscheid B, Beule D, Paolicelli RC, Meyer U. Enzymatic Dissociation Induces Transcriptional and Proteotype Bias in Brain Cell Populations. Int J Mol Sci. 2020 Oct 26;21(21):7944. doi: 10.3390/ijms21217944. PMID: 33114694; PMCID: PMC7663484.
- Denisenko, E., Guo, B.B., Jones, M. et al. Systematic assessment of tissue dissociation and storage biases in single-cell and single-nucleus RNA-seq workflows. Genome Biol 21, 130 (2020). https://doi.org/10.1186/s13059-020-02048-6