Despite cardiovascular disease being the leading cause of death worldwide and costing the US health care system almost $240 billion annually, clear diagnostic markers of disease and disease progression are lacking (1). In order to understand heart function and failure, a catalog of known cellular subtypes of the heart and deviations in their expression may provide insight into earlier diagnostic markers of heart disease or failure. Many studies have attempted to characterize heart genotype by bulk tissue sequencing, but these approaches yield results biased towards more frequently occurring subpopulations (2). Alternatively, flow cytometry and single cell RNA sequencing (scRNA-seq) have allowed researchers to identify cellular subpopulations and characterize expression levels of markers endogenous to a healthy human heart (3,4). Full characterization of diseased heart specimens, however, has been limited by challenges in obtaining high quality single cell suspensions that have not been exposed to external perturbations (2,5,6).
Here, we demonstrate the use of our Bulk Lateral Ultrasound (BLU) dissociation technology in generating high-fidelity single cell suspensions from healthy and diseased murine heart tissue with significantly reduced impact on cell markers and subpopulations relative to conventionally used approaches. BLU implements gentle dissociation with acoustic energy while maintaining samples at constant temperatures of 6°C, a fast and automated process which enables tissue dissociation into single cell suspensions in as little as 3 minutes.
To assess whether our BLU single cell dissociation technology could elucidate these differences both in diseased and healthy mice, two independent experiments were performed at Grand Valley State University through our academic collaboration with Dr. Kristin Renkema’s laboratory. The experiment contained a population (E1, N = 5) of C57BL/6 mice infected with B16 melanoma tumors for 16 days prior to harvest and a population (E2, N = 5) of mice commercially purchased. Heart tissue was harvested from both populations and dissociated using either BLU or a commercially available multi tissue dissociation kit according to the manufacturer’s protocol for the application to heart tissue. Single cell suspensions were then assessed by flow cytometry for several cell surface markers and for cell counts using CountBright™ absolute counting beads. Marker expression was determined by FlowJo and two-sample t-tests were performed in GraphPad Prism to determine significance of differences in means between conditions.
In both experiments, BLU effectively isolated a comparable number of live cells per milligram of tissue tested to those isolated using enzymes and heat (Fig. 1). Of those live cells, heart tissue dissociated by BLU yielded significantly (CD3: p-value < 0.0001 (E1) and p-value = 0.0016 (E2), CD8: p-value < 0.0001 (E1) and p-value = 0.0211 (E2)) more T cells and cytotoxic T cells than were observed with enzyme and heat dissociation (Fig. 2).
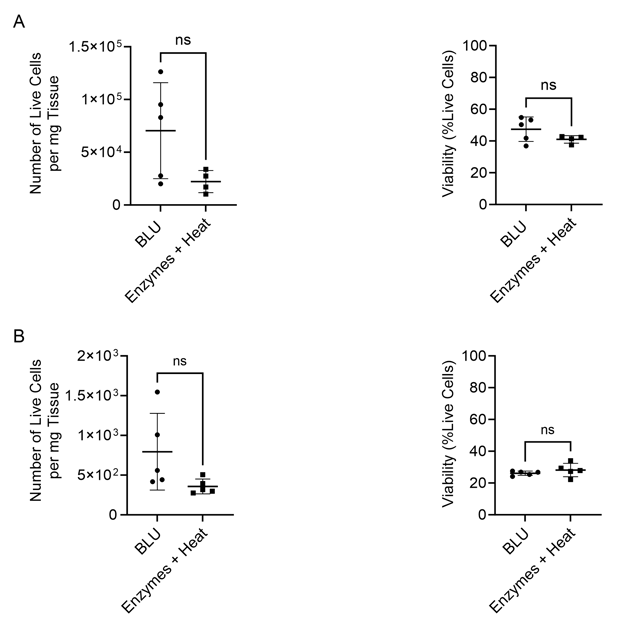
Figure 1. Comparison of cell count per milligram of tissue and total cell viability following dissociation via enzymatic or BLU approaches on heart tissue harvested from (A) C57BL/6 mice infected with B16 melanoma (E1, N = 5) or (B) mice commercially purchased (N = 5). ns = not significant
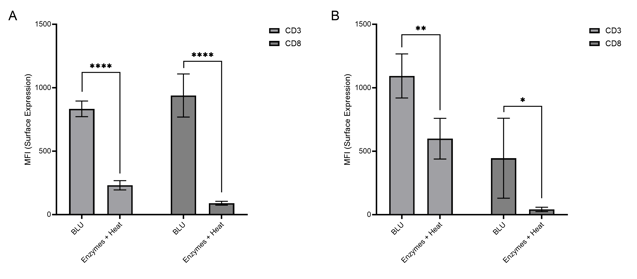
Figure 2. Comparison of CD3 and CD8 expression on live cells isolated via enzymatic or BLU dissociation of heart tissue harvested from (A) C57BL/6 mice infected with B16 melanoma (E1, N = 5) or (B) mice commercially purchased (N = 5). ns = not significant, * = p-value ≤ 0.05, ** = p-value ≤ 0.01, *** = p-value ≤ 0.001, **** = p-value ≤ 0.0001.
Further, higher levels of expression of markers for lymphocytes (CD45), macrophages (Ly6C, CD206), antigen-presenting cells (MHC-II), dendritic cells (CD86, CD11c), endothelial cells (CD144), and myelinogenic progenitor cells (CD140a) were observed on cells isolated by BLU (Fig. 3). Levels of CD11b were found to be higher on cells isolated by enzymes and heat relative to those isolated by BLU, but previous research has demonstrated an artificial upregulation of this marker during dissociation with enzymes and heat (7). These findings are consistent with previous comparisons of mouse tumor and brain tissue.
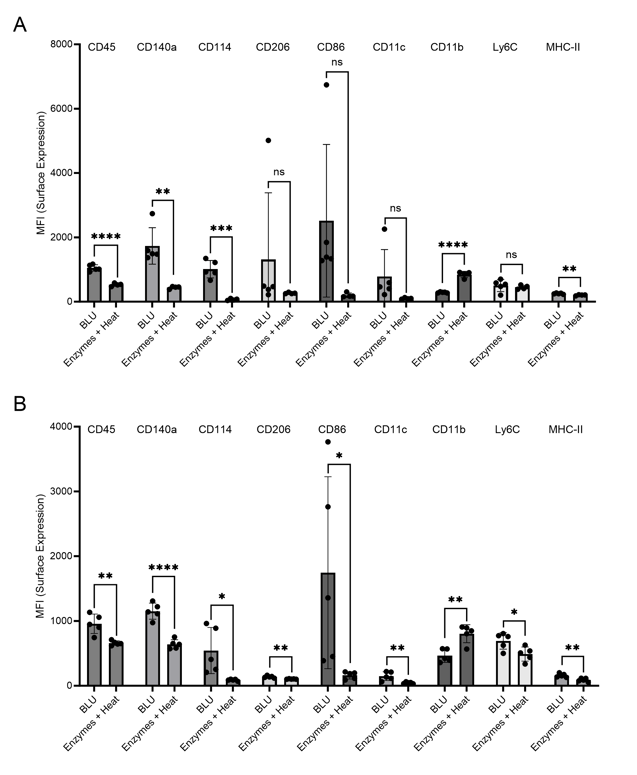
Figure 3. Comparison of myeloid cell marker expression on live cells isolated via enzymatic or BLU dissociation of heart tissue harvested from (A) C57BL/6 mice infected with B16 melanoma (E1, N = 5) or (B) mice commercially purchased (N = 5). ns = not significant, * = p-value ≤ 0.05, ** = p-value ≤ 0.01, *** = p-value ≤ 0.001, **** = p-value ≤ 0.0001.
Considering that inflammation is one of the main drivers of heart failure (4), an accurate and quantifiable understanding of immune cell infiltration is essential for characterizing disease. The inadequacy of currently established methods to preserve these immune cell subsets during heart dissociation suggests that previous studies which sought to identify markers of heart disease by flow cytometry and single cell sequencing approaches may have been deficient. The BLU technology enables enzyme and heat-free mouse heart dissociation without impacting these rare and important populations, and we highly recommend that this technology be used for obtaining higher quality single cell data.
Interested in enhancing your cell analysis with improved tissue dissociation? Ask for a demo and we will bring the SimpleFlow to you. Your cells deserve the best.
References
1. National Center for Chronic Disease Prevention and Health Promotion, Division for Heart Disease and Stroke Prevention. Center for Disease Control. 2023 [cited 2023 Aug 2]. Heart Disease Facts. Available from: https://www.cdc.gov/heartdisease/facts.htm#print
2. Pimpalwar N, Czuba T, Smith ML, Nilsson J, Gidlöf O, Smith JG. Methods for isolation and transcriptional profiling of individual cells from the human heart. Heliyon. 2020 Dec 1;6(12).
3. Litviňuková M, Talavera-López C, Maatz H, Reichart D, Worth CL, Lindberg EL, et al. Cells of the adult human heart. Nature. 2020 Dec 17;588(7838):466–72.
4. Martini E, Kunderfranco P, Peano C, Carullo P, Cremonesi M, Schorn T, et al. Single-Cell Sequencing of Mouse Heart Immune Infiltrate in Pressure Overload-Driven Heart Failure Reveals Extent of Immune Activation. Circulation. 2019 Dec 17;140(25):2089–107.
5. Koenig AL, Shchukina I, Amrute J, Andhey PS, Zaitsev K, Lai L, et al. Single-cell transcriptomics reveals cell-type-specific diversification in human heart failure. Nature Cardiovascular Research. 2022 Mar 16;1(3):263–80.
6. Gladka MM, Molenaar B, De Ruiter H, Van Der Elst S, Tsui H, Versteeg D, et al. Single-Cell Sequencing of the Healthy and Diseased Heart Reveals Cytoskeleton-Associated Protein 4 as a New Modulator of Fibroblasts Activation. Circulation. 2018;138(2):166–80.
7. Mattei D, Ivanov A, van Oostrum M, Pantelyushin S, Richetto J, Mueller F, et al. Enzymatic dissociation induces transcriptional and proteotype bias in brain cell populations. Int J Mol Sci. 2020 Nov 1;21(21):1–20.
Interested in learning more?
Ask for a 20-minute seminar from a Cellsonics Scientist




.png?width=300&height=54&name=CellsonicsLogoWhite(2048%20x%20366%20px).png)