Circulome: Single-cell morphological characterization of non-native cells circulating in the blood, a method for early detection of metastasis and non-invasive biopsies
This work was performed as part of the Cellsonics early-access program and all experiments were conducted at Dr. Euan Ashley's laboratory at Stanford University.
Saskia Botha, John E. Gorzynski, Qianru Wang, Mehdi Baqri, Euan A. Ashley
Over 90% of cancer deaths are caused by metastatic cancer1. Through hematogenous spread, tumor cells acquire the ability to invade into blood vessels and circulate through the body, sometimes resulting in tumor metastasis. Early detection of circulating tumor cells through characterizing non-native cell types in circulation may help guide treatment and reduce the risk of metastasis. Invasive biopsies are necessary to diagnose cancer and other conditions such as infections, inflammatory, and autoimmune disorders. We hypothesized that high-throughput morphological phenotyping of single-cells in the bloodstream would enable efficient identification of single-cells based on their native tissues. This would allow applications such as metastasis detection and cell culturing for non-invasive biopsies.
We optimized protocols to non-enzymatically dissociate various tissue types into single-cell suspensions using the Cellsonics SimpleFlow™ platform with Bulk Lateral Ultrasound (BLU™) energy. We harvested heart, lung, liver, spleen, kidney, brain, and muscle tissue from 10 mice. We dissociated all the tissues under different frozen conditions (fresh, overnight 4°C, flash frozen +/-80°C) using BLU energy. We then used a hemocytometer to perform cell counting and viability measurements. If viabilities were below 50%, we ran them with increasing amounts of BLU energy to find what would yield the highest viability. After optimization, we performed high-throughput imaging of single-cells from heart, liver, lung, and brain mouse tissues using the Deepcell® technology.
Figure 1. Tissues were non-enzymatically dissociated into single cell suspension with Cellsonics technology. Then, cells were imaged using the Deepcell technology or a microfluidic chip.
We extracted 100+ morphological features from the single-cell images. This study implements multi-dimensional analysis and is high-throughput which distinguishes it from previous analysis of cell morphology. Using principal component analysis, we determined the five most differentiating features for cells in each tissue type.
Uniform Manifold Approximation and Projection (UMAP) clustering of the morphological data from those 4 organs suggest that morphology is a robust method to identify cell type. Over 50% of each of the 4 clusters were specifically enriched for a single organ type.
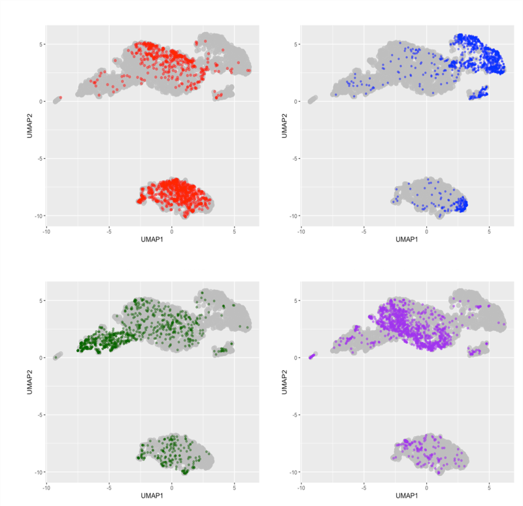
Figure 2. Heart (a), liver (b), brain (c), lung (d) cell distribution on UMAP after Deepcell image analysis. Tissue cells show ability to cluster based on morphology. Percent of each cluster that is made up of a certain organ. >50% of each cluster is from a single distinct organ.
Imperfect clustering could indicate the presence of debris and red blood cells. Using debris and dead cell removal kits may improve the accuracy of cluster analysis by reducing non-cell overlap in the morphological space.
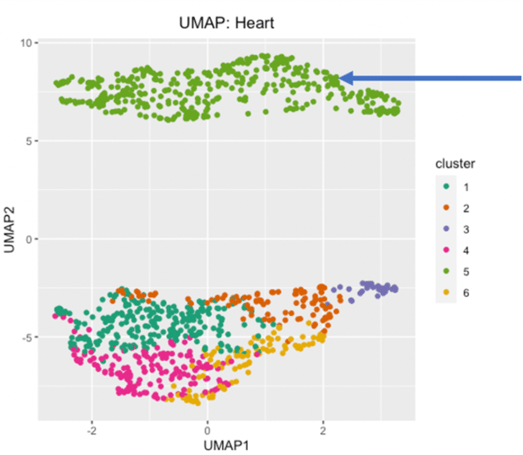
Figure 3. Clustering of cell subpopulations in heart tissue. Elbow graph of inertia per cluster (a) indicates 6-8 clusters are optimal. UMAP (b) shows 6 clusters. Cluster 1 is a distinct cell type, clusters 2-6 may consist of other subtypes or debris.
This analysis serves as a strong proof of concept that circulating cells can be classified. Cells from different tissue types are morphologically distinct enough that they can be clustered. Future directions include spiking blood with tissue cells at different concentrations to determine the sensitivity of the tissue cell classifier and using extracted morphological data beyond tissue type as a diagnostic tool.
Imaging patient blood to analyze circulating cell types will further improve personalized and precision medicine. Classification of non-native circulating cells will be helpful in identifying localized cancer by non-invasive biopsy, tumor metastasis, cell cycle state, drug responsiveness, gene expression.
References
- Fares, J., Fares, M.Y., Khachfe, H.H. et al. Molecular principles of metastasis: a hallmark of cancer revisited. Sig Transduct Target Ther 5, 28 (2020). https://doi.org/10.1038/s41392-020-0134-x
- Salek, M., Li, N., Chou, H.-P. et al. Realtime morphological characterization and sorting of unlabeled viable cells using deep learning. bioRxiv. https://www.biorxiv.org/content/10.1101/2022.02.28.482368v2.
Interested in learning more?
Ask for a 20-minute seminar from a Cellsonics Scientist



.png?width=300&height=54&name=CellsonicsLogoWhite(2048%20x%20366%20px).png)