As researchers continue to map the architecture of the brain and identify rare subsets of cells with essential functions, single cell analyses of brain tissue are becoming increasingly important. Many researchers rely on tissue dissociation to prepare single cell suspensions for downstream multiomic applications (1). The conventional approach for tissue dissociation involves treatment with enzymes and sometimes heat, a process which Mattei et al have demonstrated introduces significant transcriptional and proteotype bias in bran cells (2).
Having previously demonstrated enhanced CD8+ T cell dissociation from B16 melanoma tumors collected from C57BL/6 mice, we* wanted to compare our SimpleFlow™ acoustic dissociation technology (BLU) to conventional enzyme and heat-based dissociation of brain tissue. SimpleFlow implements gentle dissociation with acoustic energy while maintaining samples at constant temperatures of 6°C, a fast and automated process which enables tissue dissociation into single cell suspensions in as little as 3 minutes.
To assess whether our SimpleFlow acoustic based single cell dissociation technology could elucidate these differences both, two independent experiments were performed at Grand Valley State University through our academic collaboration with Dr. Kristin Renkema’s laboratory. In these experiments where B16 melanoma tumors had been allowed to grow for 12-16 days, brains were collected post-harvest to yield three biological replicates per condition (N = 3) in the first experiment (E1) and five biological replicates per condition (N = 5) in a second follow-up experiment (E2). Brain tissue was dissociated using SimpleFlow or a commercially available brain tissue dissociation kit according to the manufacturer’s protocol. Single cell suspensions were then assessed by flow cytometry for several cell surface markers and for cell counts using CountBright™ absolute counting beads. Marker expression was determined by FlowJo and two-sample t-tests were performed in GraphPad Prism to determine significance of differences in means between conditions.
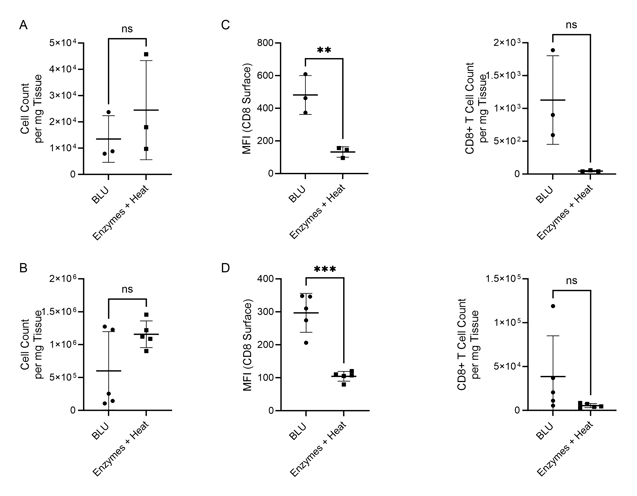
Figure 1. Comparison of cell count per milligram of tissue, CD8 surface expression and CD8+ T cell count per mg of tissue following B16 melanoma tumor dissociation by enzymatic or BLU approaches. (A) E1 cell count per mg tissue, N = 3 biological replicates per group. (B) E2 cell count per mg tissue, N = 5 biological replicates per group. (C) E1 (N = 3) CD8 surface expression and CD8+ T cell subset counts per milligram of tissue. (D) E2 (N = 5) CD8 surface expression and CD8+ T cell subset counts per milligram of tissue. ns = not significant, ** = p-value ≤ 0.01, *** = p-value ≤ 0.001.
Figure 2. Co-expression of CD45 and CD3 on CD8+ T cells isolated by BLU tissue dissociation in E1 (N = 3). (A) CD8+ T cell selection. (B) CD3 and CD45 co-expression on BLU dissociated CD8+ T cell subsets. Too few CD8+ T cells were isolated by enzymatic dissociation to enable a comparable assessment.
Because these mice had been developing tumors for 12-16 days, we wanted to investigate inflammation of the brain and thus looked for markers of leukocyte (CD45, CD3, CD4, and CD8) and myeloid cell infiltration (CD115, CD11b, Ly6C, Ly6G, CD86, F4/80, and CD206), similarly to analyses performed by Burfeind et. al. (3).
While in both experiments the total number of cells dissociated by BLU or enzymes were not significantly different (Fig. 1A & B), we demonstrated that the CD8 expression of cells isolated using BLU was significantly (p-value = 0.0081 in E1, p-value ≤ 0.001 in E2) higher than the CD8 expression of T cells isolated by enzymes (Fig. 1C & D). This led to a much larger population of CD8+ T cells to run further studies on which were not possible with the enzymes and heat-dissociated population due to their prohibitively low numbers in both E1 and E2 (Fig. 2A & Fig. 3A). Wanting to further understand the phenotype of the CD8+ T cells isolated from the brain tissue, we demonstrated CD3 and CD45 expression on this subset from both E1 and E2 relative to cells which were negative for CD4 and CD8 expression (Fig. 2B & Fig. 3B).
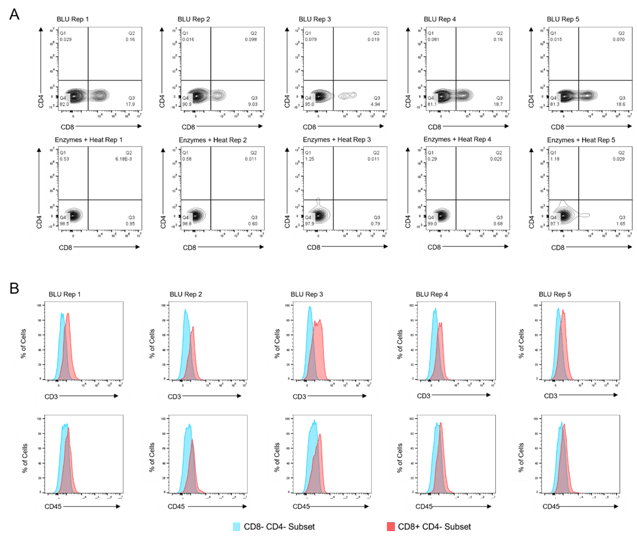
Figure 3. Co-expression of CD45 and CD3 on CD8+ T cells isolated by BLU tissue dissociation in E2 (N = 5). (A) CD8+ T cell selection. (B) CD3 and CD45 co-expression on BLU dissociated CD8+ T cell subsets. Too few CD8+ T cells were isolated by enzymatic dissociation to enable a comparable assessment.
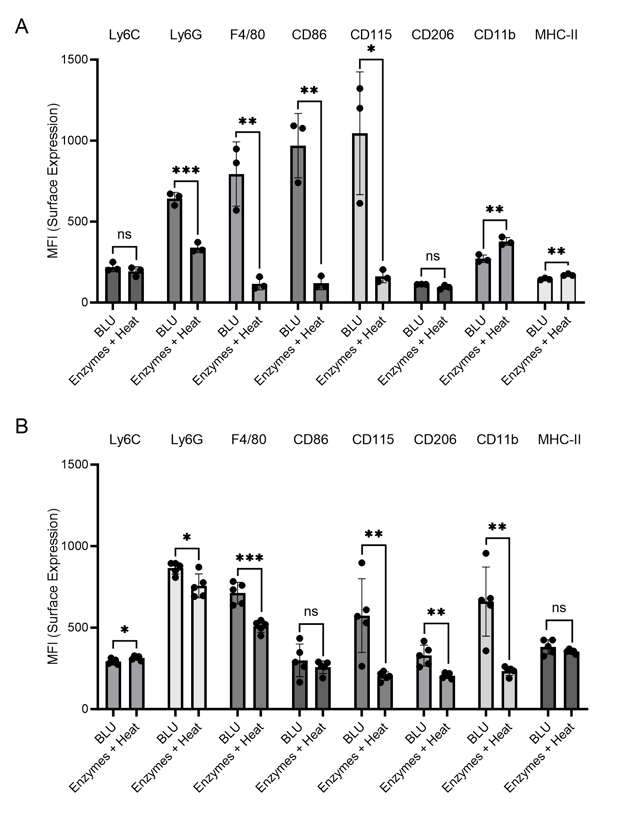
Figure 4. Cell surface expression (MFI) of myeloid cell markers on cells dissociated from brain tissue by BLU or enzymes and heat. (A) N = 3 biological replicates per group. (B) N = 5 biological replicates per group. ns = not significant, * = p-value ≤ 0.05, ** = p-value ≤ 0.01, *** = p-value ≤ 0.001.
Looking further at the myeloid cell markers, we identified in both experiments significantly higher expression of Ly6G, F4/80, CD86, and CD115 on cells isolated from brain tissue by BLU relative to those isolated by enzymes and heat (Fig. 4). Especially during brain inflammation, which we expected for the tumor-ridden mice in these experiments, it is often difficult to distinguish between brain-resident macrophages, or microglia, and inflammatory monocyte-derived cells (4). While F4/80 and CD115 are expressed on both subtypes, Greter, et. al. suggest that CD86 may help differentiate activated microglia, making it essential to be able to characterize this marker. Conversely, Ly6G has been previously identified on brain-infiltrating neutrophils in a mouse model of pancreatic ductal adenocarcinoma, making this marker a distinct indicator of infiltrating myeloid cells (3). The loss of expression of these markers due to the use of enzymes and heat for tissue dissociation directly impacts researchers’ ability to differentiate cell subtypes and understand activities in the brain during inflammation.
One marker was found to be upregulated in samples dissociated using enzymes and heat relative to BLU, CD11b. This commonly used microglial marker was previously found to be artificially upregulated by enzymes and heat relative to mechanical dissociation, suggesting a concerning bias arising from this type of tissue dissociation (2).
In this study, we have demonstrated the capacity of the SimpleFlow™ BLU technology to not only dissociate more cells of interest from brain tissue than conventional methods employing enzymes and heat, but also to more consistently and dependably maintain cell surface expression of important cellular subsets. As neural research continues developing, this technology will be essential in elucidating rare subpopulations and enabling studies of cells with higher integrity, and should be strongly considered as a replacement for enzymes and heat for researchers working with brain tissue.
Interested in enhancing your cell analysis with improved tissue dissociation? Ask for a demo and we will bring the SimpleFlow to you. Your cells deserve the best.
References:
1. G. La Manno, K. Siletti, A. Furlan, D. Gyllborg, E. Vinsland, A. M. Albiach, C. M. Langseth, I. Khven, A. R. Lederer, L. M. Dratva, A. Johnsson, M. Nilsson, P. Lonnerberg and S. Linnarsson, "Molecular architecture of the developing mouse brain," Nature, vol. 596, 2021.
2. D. Mattei, A. Ivanov, M. van Oostrum, S. Pantelyushin, J. Richetto, F. Mueller, M. Beffinger, L. Schellhammer, J. vom Berg, B. Wollscheid, D. Beule, R. C. Paolicelli and U. Meyer, "Enzymatic Dissociation Induces Transcriptional and Proteotype Bias in Brain Cell Populations," International Journal of Molecular Sciences, vol. 21, 2020.
3. K. G. Burfeind, X. Zhu, M. A. Norgard, P. R. Levasseur, C. Huisman, A. C. Buenafe, B. Olson, K. A. Michaelis, E. R. Torres, S. Jeng, S. McWeeney, J. Raber and D. L. Marks, "Circulating myeloid cells invade the central nervous system to mediate cachexia during pancreatic cancer," eLife, 2020.
4. M. Greter, I. Lelios and A. L. Croxford, "Microglia versus myeloid cell nomenclature during brain inflammation," Frontiers in Immunology, 2015.
Interested in learning more?
Ask for a 20-minute seminar from a Cellsonics Scientist




.png?width=300&height=54&name=CellsonicsLogoWhite(2048%20x%20366%20px).png)